Kate and Anton
Kate and Anton are a match made not in heaven but at the Pittsburgh Supercomputing Center. Kate is Kate Stafford, a fifth-year National Science Foundation predoctoral fellow in my laboratory in the Department of Biochemistry & Molecular Biophysics. Anton is a supercomputer specialized to perform molecular dynamics—MD—simulations of proteins and other biological macromolecules.
Anton was developed at D.E. Shaw Research, a Manhattan-based research institute headed by David E. Shaw, PhD (who is also senior research fellow at the Center for Computational Biology and Bioinformatics, or C2B2, at Columbia). Anton is made available to the research community through a collaboration between the Pittsburgh Supercomputing Center and the Shaw institute with support from the NIH National Institute of General Medical Sciences.
Ms. Stafford uses Anton in her doctoral research through a grant titled “MD and NMR characterizations of psychrotrophic, mesophilic, and thermophilic enzymes” from the Pittsburgh Supercomputing Center.
Proteins perform myriad functions in biology, ranging from structural roles, to recognition and transport of other molecules, to catalysis of chemical reactions. Proteins are essential to infectivity and virulence of pathogens. Mutations, misfolding, and other malfunctions of proteins are implicated in numerous human diseases so it is not surprising that proteins are the targets of most pharmaceutical agents and emerging treatments, such as gene therapy. Paradoxically, protein function depends upon two opposed constraints: The molecules must be sufficiently stable to maintain their active 3-D structures but sufficiently flexible to perform their biological and chemical actions. The interplay between stability and flexibility is exemplified by proteins isolated from bacteria adapted to live at very different temperatures, because atomic motions and, hence, flexibility of molecules increase with increasing temperature. Psychrotrophic, mesophilic, and thermophilic bacteria have been identified that thrive at low (in some cases less than 0 degrees C), moderate, and high (in some cases more than 100 C) temperatures. Proteins isolated from thermophilic bacteria frequently are more stable but less biologically active at ambient temperatures. In contrast, proteins isolated from mesophilic bacteria lose activity at elevated temperatures owing to disruption of the proper 3-D structures.
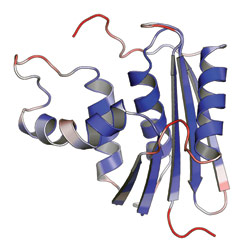 Amplitudes of motion calculated from an MD simulation are mapped onto the 3-D structure of E. coli RNaseH. Blue and red regions indicate low and high local flexibility, respectively. Elements of secondary structure, α-helices and β-sheets, are rigid, while peripheral loops, which are known to participate in binding the substrate and carrying out the protein’s catalytic reaction, are more flexible.
Amplitudes of motion calculated from an MD simulation are mapped onto the 3-D structure of E. coli RNaseH. Blue and red regions indicate low and high local flexibility, respectively. Elements of secondary structure, α-helices and β-sheets, are rigid, while peripheral loops, which are known to participate in binding the substrate and carrying out the protein’s catalytic reaction, are more flexible.Ms. Stafford’s research is directed toward a fundamental understanding of how stability and flexibility of proteins are balanced to achieve the requisite biological activity. She has focused on the protein ribonuclease HI (RNaseH), an enzyme that is widespread in prokaryotes and eukaryotes (and also is found as a domain of the reverse transcriptase enzyme of retroviruses). The enzyme degrades the RNA strand of DNA:RNA hybrid molecules and is involved in numerous activities related to replication of DNA. RNaseH also is critical to efforts to develop “antisense” biotechnology in which specific RNA molecules are targeted for destruction. Ms. Stafford has chosen RNaseH homologs from the bacteria Shewanella oneidensis (soRNH), Eschericia coli (ecRNH), and Thermus thermophilus (ttRNH) for comparative investigations of the properties of the enzymes from organisms capable of growth from less than 10 C to more than 60 C.
My laboratory has used NMR spectroscopy to experimentally characterize the conformational flexibility of RNaseH enzymes over multiple time scales. However, distinguishing between different mechanisms of motion, which is critical for understanding biological function, is difficult using experimental data alone. MD simulations, provided that they are accurate over time scales comparable to the experimentally relevant ones, enable deep insights into mechanism. Hence, joint analysis of experimental NMR spectroscopy and MD simulations is a powerful approach to understanding functions of proteins (or other biomolecules). Previously, former graduate student Nikola Trbovic and Ms. Stafford performed extensive MD simulations of all three homologs for times up to 100 ns-1 ms over a temperature range from 0 C to 67 C. These MD simulations have been performed on computer clusters in my laboratory and at C2B2 and represent the state of the art for conventional computer hardware. Nonetheless, the lengths of the MD simulations are still much less than necessary to allow full comparisons with the experiments.
Using Anton, Ms. Stafford is extending the length of the MD simulations of these proteins by one-to-two orders of magnitude. An example of some of the initial results for ecRNH are shown in the illustration that depicts the relative amplitudes of motion mapped onto a representation of the protein structure. Already, these simulations are suggesting ways in which the three proteins respond differentially to changes in temperature and are targeting amino acid residues critical for function. Ms. Stafford and other members of my group will then produce the mutated proteins and repeat the NMR experiments to test the predictions from the simulations. Ultimately, Ms. Stafford’s work with Anton will provide an atomically detailed description of how one family of proteins has adapted to function in the diverse environments in which the host organisms live and, we hope, provide more general insights into the remarkable properties of proteins essential for life.
Kate and Anton illustrate a fundamental feature of biological, indeed all, science: New technologies drive new areas of research while quantitative improvements of technology eventually lead to qualitative transformations of applications. More than three centuries ago, Anton Van Leeuwenhoek revealed a dynamic sphere of microscopic bacteria and today his namesake computer reveals the dynamic behavior of their constituent molecules.
- Log in to post comments


